Targeted therapy has undoubtedly transformed the treatment landscape for metastatic NSCLC, with multiple agents now available to target seven different molecular drivers. In tandem with the growing treatment armamentarium, comprehensive molecular profiling has been critical in providing insight into the complexity of a patient’s disease and enabling precision treatment.
“Tremendous progress has been made, and I think we are really improving our ability to deliver targeted therapy based on improvements in liquid biopsy,” according to Charu Aggarwal, MD, MPH, the Leslye M. Heisler Associate Professor for Lung Cancer Excellence at the University of Pennsylvania’s Perelman School of Medicine.
Despite these advances, all targeted agents face a similar consequence. “After receipt of targeted therapies, emergence of resistance is unfortunately inevitable and something that we must be prepared for,” Dr. Aggarwal cautioned.
Some resistance mutations are well established and highly predictable, such as emergence of the EGFR T790M mutation that commonly develops during treatment with first-generation EGFRdirected tyrosine kinase inhibitors. Clinicians often know how to deal with these common mutations—in this case, by administering osimertinib, a thirdgeneration EGFR inhibitor with activity against T790M mutation-positive NSCLC. But what happens when the patterns of resistance have not yet been defined or are highly variable, and treatment decisions need to be made quickly to address disease progression? In these situations, Dr. Aggarwal feels that “ctDNA is a very valuable tool for picking up a resistance mechanism that is immediately actionable.”
Use in Therapeutic Selection

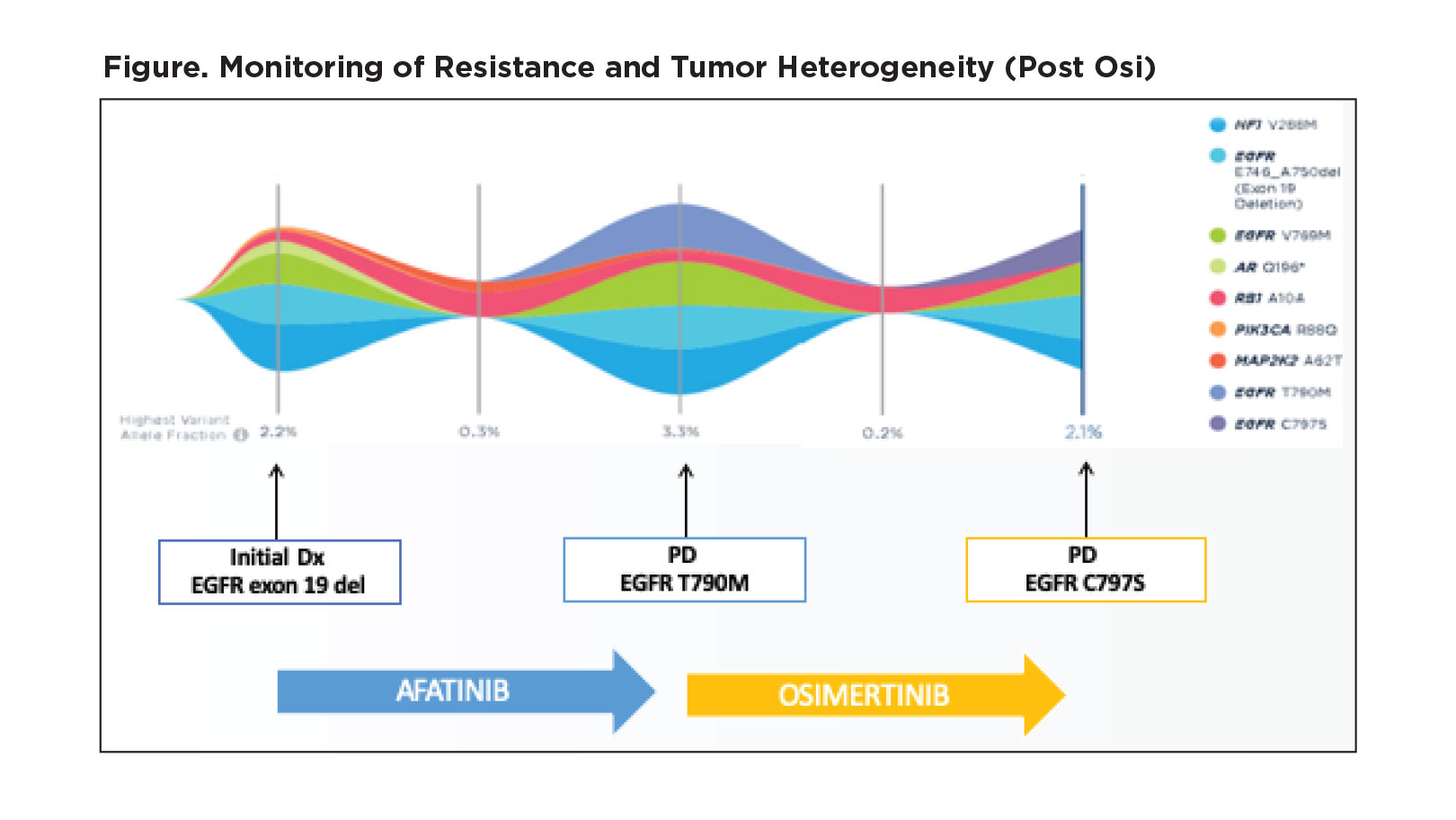
Dr. Aggarwal provided several examples in which ctDNA can be used to identify the emergence of resistant mutations and, in turn, steer treatment selection
for a patient. For example, she shared details about one of her patients with an EGFR exon 19 deletion who she initially treated with afatinib (Figure). Upon development of radiographic progression, the patient was found to have an EGFR T790M mutation by ctDNA analysis, and she switched the patient to osimertinib. The patient later developed clinical and radiographic signs of progression, and ctDNA testing revealed the presence of the EGFR C797S mutation. At that point, a clinical trial offered the best option for treatment.
In other cases, ALK fusions have been detected in ctDNA as a mechanism of resistance to osimertinib, prompting use of combination therapy with osimertinib and an ALK tyrosine kinase inhibitor. Of two individuals found to have EGFR L858R and an ALK fusion that Dr. Aggarwal showed as examples, one received osimertinib and crizotinib and achieved stable disease following just one cycle of combination therapy, and the other received osimertinib and alectinib and showed a 25% reduction in the dominant mass after one cycle.1
Serial ctDNA testing also lends insight into the longitudinal development of compound mutations within a single gene. For example, research shows that ALK resistance mutations accumulate with successive lines of treatment, particularly when patients relapse on lorlatinib.2 Th e most frequent combinations of ALK mutations observed consist of G1202R plus L1196M and D1203N plus I1171N. Although these mutations may not be clinically actionable at this time, Dr. Aggarwal stated that this research demonstrates that “we can overcome the problem of tissue heterogeneity by capturing the global mutational paradigm by assessing plasma.” To incorporate ctDNA in the clinic to aid in sequencing targeted therapy, Dr. Aggarwal says that the first step following diagnosis and molecular profiling is to choose the appropriate targeted therapy in a personalized manner for the patient. “At the time of progression or development of treatment resistance, ctDNA can then be used to inform either singleagent therapy or combination therapy. As I discussed with you, in previous reports, there have been combinations of different targeted therapies leading to clinical and radiographic improvement” she said.
Despite the utility of ctDNA for rapidly elucidating resistance mechanisms, Dr. Aggarwal warned, “We must still be cognizant of transformation events that require tissue biopsy.” Th is includes cases of small cell transformation in patients with EGFR-mutated or ALK-translocated disease, as well as possible transformation to squamous cell histology.
References
1. Offi n M, Somwar R, Rekhtman N, et al. Acquired ALK and RET Gene Fusions as Mechanisms of Resistance to Osimertinib in EGFR-Mutant Lung Cancers. JCO Precis Oncol. 2018;2:PO.18.00126.
2. Dagogo-Jack I, Rooney M, Lin JJ, et al. Treatment with next-generation ALK inhibitors fuels plasma ALK mutation diversity. Clin Cancer Res. 2019;25(22):6662-6670.