
Non-small cell lung cancer (NSCLC) is a heterogeneous disease with various subgroups that may benefit from different treatments. Commonly, NSCLC is divided into smoking-associated and non-smoking associated tumors based on patient-reported smoking history to aid in predicting in which subgroup a patient might fall. This distinction is important as the molecular makeup of lung cancer can differ greatly between patients who have never smoked and those who have been exposed to tobacco smoke.1,2,3
However, relying solely on patient-reported smoking history may not accurately reflect the true molecular landscape of the tumor, as non-smoking-related oncogenic mutations can also occur in patients who smoke or have previously smoked. Therefore, current guidelines recommend testing all adenocarcinomas for driver mutations regardless of smoking history.4 Thus, clinical smoking history has limited diagnostic or therapeutic consequences in daily clinical practice. In contrast, the clinical trials that guide daily practice still frequently use patient-reported clinical smoking history as stratification factor or for subgroup analyses.
The ever-expanding landscape of molecular diagnostics in NSCLC provides an opportunity to more accurately classify tumors as smoking-associated and non-smoking associated. For instance, next generation sequencing (NGS) can be used to identify patterns of somatic mutations, known as mutational signatures, that are caused by various endogenous and exogenous processes.5 Some of these signatures have previously been associated with tobacco smoking and could therefore potentially allow for a classification of smoking- and non-smoking-associated NSCLC based on individual genomic tumor characteristics. Despite NGS being standard of care in most practices, a genome-based classification of NSCLC has not yet been incorporated into daily practice or clinical research.
We investigated the potential of a mutational signature-based classification rather than the current classification that is based on clinical smoking history.6 We used whole genome sequencing and clinical data from three prospective cohorts of metastatic NSCLC (n=316). We used two tobacco smoking signatures in combination with background age-related signatures to divide the cohort into two clusters: one cluster in which tobacco smoking-related mutational signatures were considerably present in the cancer genome—the ‘smoking high’ cluster—and one cluster in which they were not—the ‘smoking low’ cluster.
One of the major findings of our study was that a substantial subset of metastatic NSCLC is differently classified into smoking- and non-smoking-associated tumors based on smoking-related mutational signatures than based on smoking history (Figure 1). Although most patients who reported never smoking had little or no smoking-related signatures in their tumors, four out of 89 patients who had never smoked were still included in the smoking high cluster. Further analysis of these cases showed that they had a similar molecular profile to the other patients in the smoking high cluster. As these smoking signatures have not been associated with other endogenous or exogenous causes other than tobacco smoke exposure, it is probable that these four patients were indeed exposed to tobacco smoke, either directly or secondhand, despite their negative self-reported smoking history.
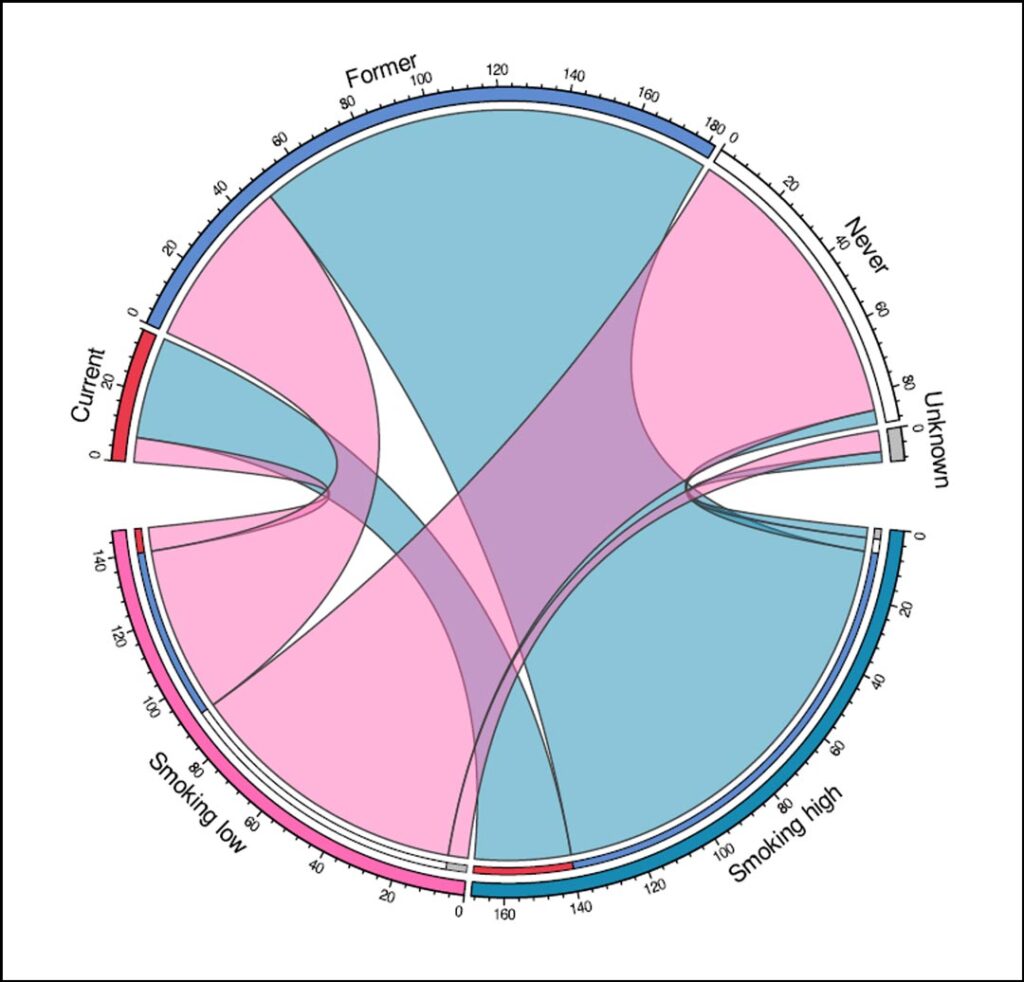

Additionally, the presence of a KRAS G12C mutation in one case and the lack of a known driver oncogene in another case, supports the likelihood of exposure to tobacco smoke. This discrepancy between genomic landscape and clinical smoking history could be due to recall bias, inaccurate history taking by healthcare professionals, or secondhand smoke exposure. These cases illustrate one of the shortcomings of clinical smoking history as there are many factors that can influence its reliability.
Besides the limited reliability of clinical smoking history, there is another limitation to its usefulness in daily practice and clinical research. We found that the amount of smoking signature contribution in patients with a history of smoking was highly variable. A total of 26% of patients who had smoked or were currently smoking did not show high smoking signature contribution. Moreover, more than half of these patients harbored a targetable oncogene that is not related to smoking (e.g. EGFR/ALK/RET/ROS1). Presumably, this clinical-genomic discrepancy is also influenced by the amount of tobacco smoke exposure.
For instance, we found that those included in our smoking low cluster had significantly fewer pack years than those in the smoking high cluster. However, the amount of pack years is rarely collected in a structured fashion in daily practice and is often only a rough estimation. Additionally, it does not take other aspects of smoking behavior into account, such as inhalation behavior and time since quitting. This demonstrates that if molecular testing is only guided by smoking history, and additional analyses, such as comprehensive RNA analysis, are therefore not pursued in patients with an active smoking history, (rarer) targetable driver oncogenes might go undetected.

The variability in smoking signature contribution in patients with a history of smoking also raises questions about the usefulness of smoking history in clinical research. Specifically, in trials investigating immunotherapy, smoking history is often used as a predictor of tumor molecular profile and potential response to immunotherapy. However, our findings suggest that using smoking history as a stratification factor may not accurately distribute patients into groups with similar molecular profiles. By using molecular smoking signatures instead, treatment effects could be more accurately evaluated within subgroups with similar molecular characteristics.
So, mutational signature analysis can have implications for daily clinical practice as well as clinical research. However, it can also have a broader impact on patients who suffer from lung cancer. Lung cancer is often viewed as a preventable and therefore self-inflicted disease, leading to feelings of guilt and self-blame in patients. This stigma can negatively affect their mental and emotional health, and even cause them to misreport their smoking history to healthcare professionals, further diminishing the reliability of clinical smoking history.7,8 Thus, reducing the stigma associated with lung cancer by removing the label of “smoker” from clinical practice and research can help reduce this burden on patients who are already facing a serious illness.
Our study is the first to focus on the discrepancies between clinical smoking history and the mutational processes in the cancer genome and highlights the limitations of relying solely on clinical smoking history. While patient-reported clinical smoking history still holds certain value in the present day, our findings suggest that tobacco smoking related mutational signatures are promising markers to potentially guide molecular testing and optimize patient stratification in trials. Of note, our study showed that these signatures can also be identified using targeted NGS panels, making it more feasible for implementation in daily clinical practice and research.
As molecular panels used in daily practice continue to evolve, mutational signatures could play an increasingly important role as biomarkers. This would allow for the identification of molecularly related tumors within the heterogeneity of NSCLC, which in turn could lead to more personalized and effective treatment options for patients with lung cancer.
References
- 1. Chapman AM, Sun KY, Ruestow P, et al. Lung cancer mutation profile of EGFR, ALK, and KRAS: Meta-analysis and comparison of never and ever smokers. Lung Cancer 2016;102:122-134.
- 2. Govindan R, Ding L, Griffith M, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 2012;150:1121-1134.
- 3. Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers–a different disease. Nat Rev Cancer 2007;7:778-790.
- 4. Planchard D, Popat S, Kerr K, et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2018;29:iv192-iv237.
- 5. Alexandrov LB, Kim J, Haradhvala NJ, et al. The repertoire of mutational signatures in human cancer. Nature 2020;578:94-101.
- 6. Ernst SM, Mankor JM, van Riet J, et al. Tobacco Smoking-Related Mutational Signatures in Classifying Smoking-Associated and Nonsmoking-Associated NSCLC. J Thorac Oncol 2022.
- 7. Hamann HA, Ver Hoeve ES, Carter-Harris L, et al. Multilevel Opportunities to Address Lung Cancer Stigma across the Cancer Control Continuum. J Thorac Oncol 2018;13:1062-1075.
- 8. Stuber J, Galea S. Who conceals their smoking status from their health care provider? Nicotine Tob Res 2009;11:303-307.





